
Technology Development
We drive our technology development through 7 translational Technology Focus Areas (TFAs) to ensure coordination within the platform as well as long-term competitive service offering to researchers, clinical trials, and healthcare at the internationl forefront in the specific technology domains.

Advanced Data Analytics
Clinical bioinformatics and AI play a crucial role in translating large-scale datasets into actionable insights for personalized medicine. We focus on:
- Optimization of current algorithms following FAIR guidelines
- Integration of diverse data types (e-records, imaging, omics) to create comprehensive patient profiles
- Evaluation and validation of AI models to ensure their reliability and relevance
- Increasing scalability in diagnostics, improving outcomes and healthcare delivery
Coordinating node: Gothenburg

Epigenomics
Recent years have witnessed the development of methylation-based classifiers for several cancer types and the CG platform has been instrumental in assessing the clinical utility of this tool for correct identification and treatment of central nervous system tumors. We focus on:
- National implementation of methylation-based classifiers for CNS tumors
- Improving diagnostic accuracy—changing diagnosis in 1 in 10 CNS tumor cases
- Developing and validating pipelines and classifiers for international clinical trials
- Expanding to cardiopulmonary and infectious disease classifiers
- Exploring real-time intraoperative tumor classification using custom panels and long-read sequencing
Coordinating node: Linköping
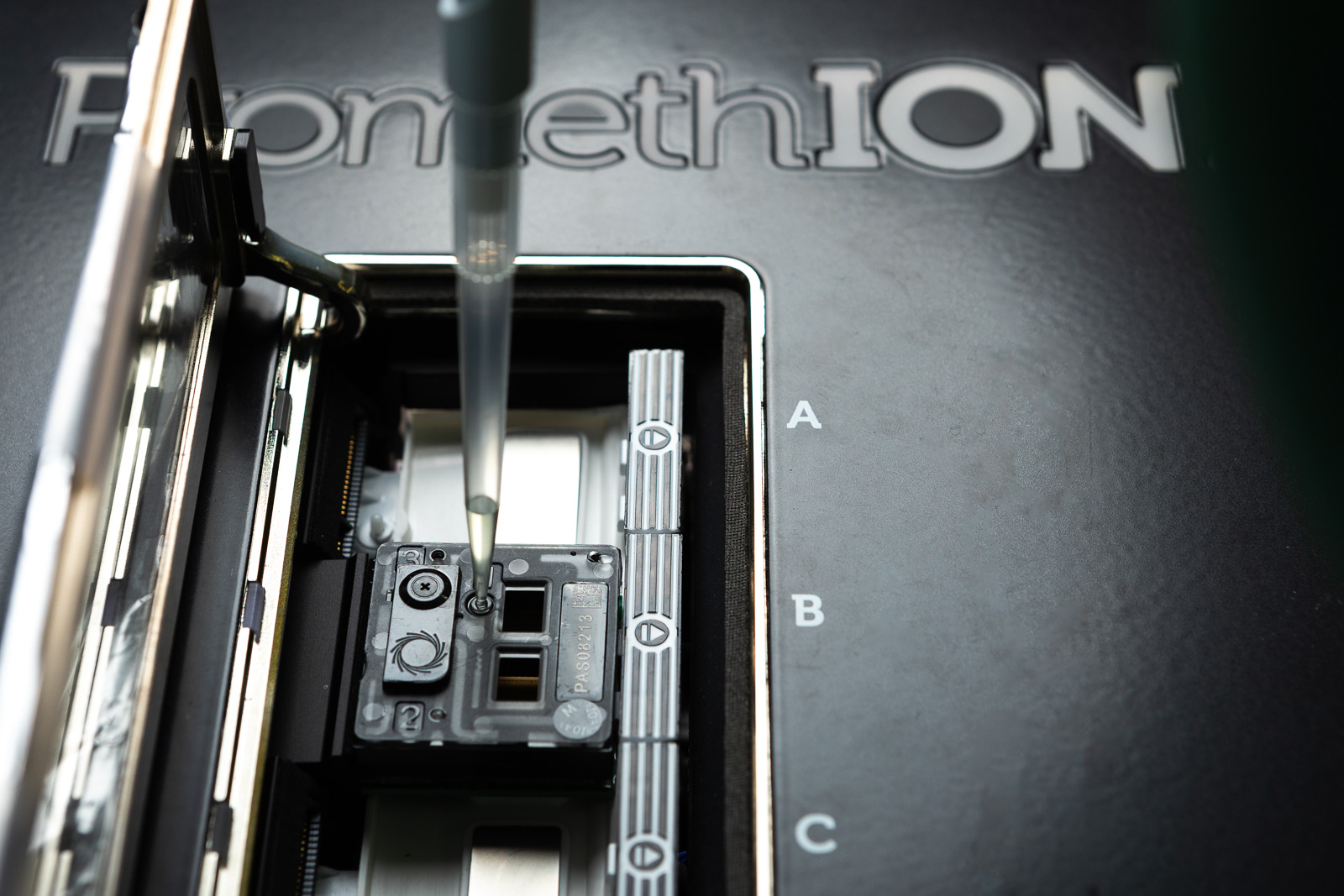
Long-read sequencing
Recent advancements in long-read sequencing technologies improve SNV, structural variant, repeat expansion, and methylation profile identification, potentially serving as a comprehensive diagnostic tool. We focus on:
- Improving rare disease diagnostic yield beyond 40% achieved by short-read WGS
- Replacing time-intensive methods in cancer diagnostics, such as chromosome analysis
- Accelerating microbial genome assembly for rapid species ID and resistance prediction
- Detecting complex structural variants using optical mapping in difficult genomic regions
- Integrating clinical assays, including SARS-CoV-2 and BCR::ABL1 fusion gene detection
Coordinating node: Uppsala
Metagenomics
Comprehensive analysis of microbial and host genetic material holds great potential for diagnosis and treatment of infectious disease. We serve as the technical backbone for several national projects and focus on:
- Clinically validated, rapid metagenomic sequencing assays for detection of known and unknown pathogens
- Human background depletion or microbial enrichment
- Antimicrobial resistance prediction
- Publicly available bioinformatic pipelines
Coordinating node: Örebro

Multi-modal omics
The addition of multiple data layers has been shown to improve diagnostic yield in several disease areas. We focus on:
- Leveraging transcriptome sequencing to detect regulatory variants, splicing defects, gene fusions, and allelic imbalances
- Refining bioinformatic workflows and integrating RNA, DNA, and methylation data into joint variant prioritization
- Investigating gene inactivation through combined expression and methylation profiling
- Exploring a joint proteogenomics diagnostic service in collaboration with the Clinical Proteomics platform
Coordinating node: Stockholm
Single-cell omics
In recent years, single-cell applications have demonstrated great potential to advance future healthcare diagnostics, prognostics, and disease monitoring. We focus on:
- Providing single-cell transcriptomics, epigenetics, and multimodal analyses via 10x Genomics and MissionBio platforms
- Increasing throughput and cost-efficiency, particularly for low-input clinical samples
- Collaborating with NGI and Spatial Biology platforms to expand into proteomics and high-resolution spatial omics (e.g., in situ sequencing)
Coordinating node: Lund
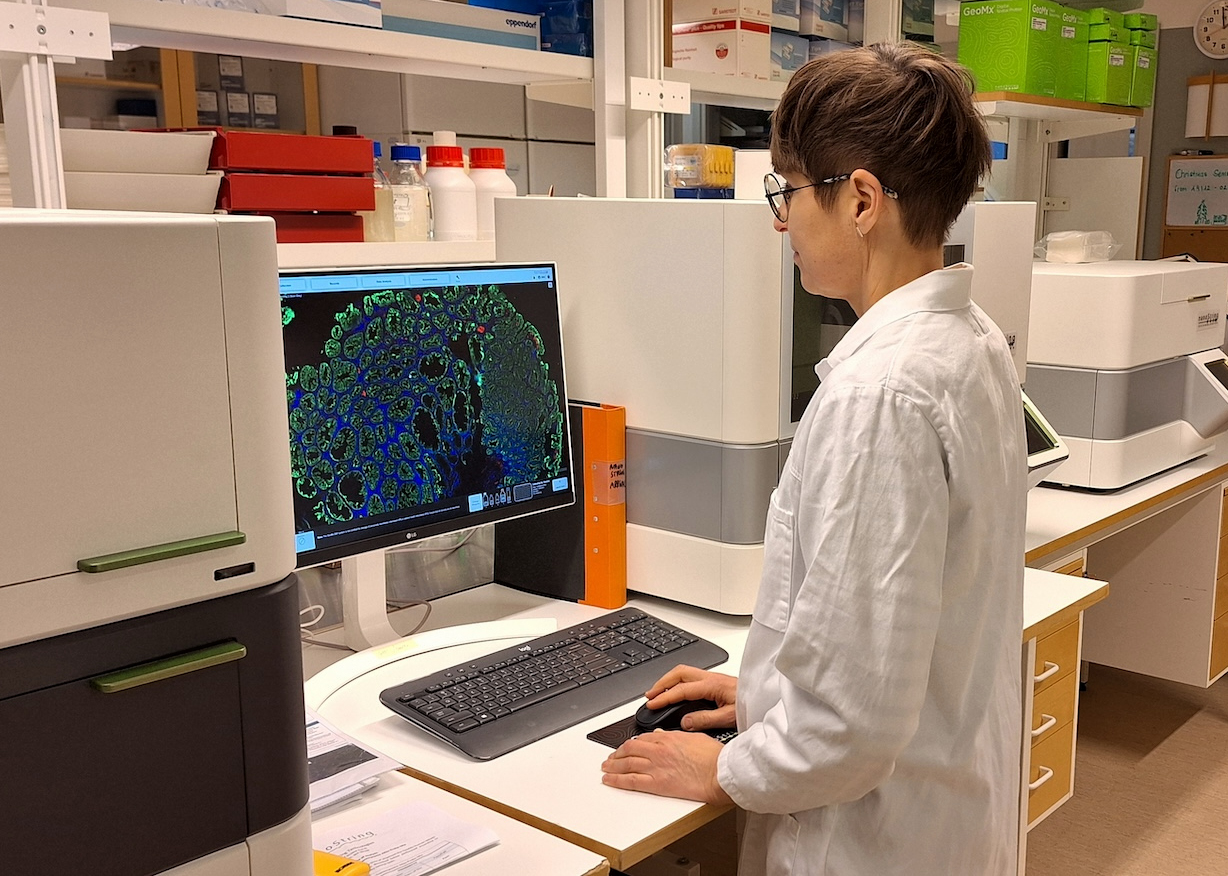
Spatial omics
Spatial omics enables high-resolution mapping of molecular features within tissues, preserving spatial context critical for understanding complex biological systems, advancing diagnostics, prognostics and therapeutic strategies. We focus on:
- Offering spatial analyses using NanoString GeoMx and 10x Genomics CytAssist
- Collaborating with NGI and Spatial Omics platforms to further expand the proteomic and high-resolution spatial omic (e.g., in situ sequencing) services
Coordinating node: Umeå
Ultrasensitive detection
Ultra-sensitive assays for identification of low frequency mutations play a crucial role in cancer diagnosis, prognosis and monitoring treatment response. We focus on:
- Combining digital PCR, deep sequencing with UMIs, and superRCA for optimal sensitivity and efficiency
- Implementing SimSen-Seq as a national multi-marker detection service
- Developing liquid biopsy-based diagnostics across multiple cancers using circulating tumor DNA
- Conducting a clinical study in MDS to predict relapse post-transplant using digital PCR and superRCA
Coordinating node: Uppsala
